
 English
English
 French
French
Study of the diversity of molds and their toxicity for systemic and respiratory health in soybean sauce
Etude sur la diversité des mycoses et leur toxicité pour le problème de santé systémique et respiratoire
H. Le Thi Viet 1, H. Lai Duy 2, T. Doan Van 3
1: Ngo Quyen High School. Le Chan District. Hai Phong city. Vietnam
2: Tran Phu Specialized High School. Hai Phong city. Viet Nam
3: Hanoi Pedagogical University. Hanoi - Viet Nam
Corresponding author
A/Professor DOAN VAN Thuoc
Hanoi Pedagogical University. Hanoi - Viet Nam
Email: doanvanthuoc@gmail.com
DOI: 10.12699/jfvpulm.9.26.2018.32
ABSTRACT
Introduction. Aflatoxins are mycotoxins secreted by Aspergillus flavus, which can colonize the respiratory tract and cause fungal rhinosinusitis or bronchopulmonary aspergillosis. A. flavus is the second leading cause of invasive aspergillosis worldwide.
Objectives. Objectives of the study was to evaluate the diversity of mold and fungal toxins in some soybean sauce samples collected in Hung Yen induced respiratory and systemic health problem.
Methods. Samples were randomly collected in some families with traditional soybean sauce production and some samples were sold on the market in Ban village - Hung Yen town. Microorganisms: fungal isolated from the soybean sauce collected in Ban village - Hung Yen town.
Results. The result showed that the moldy colonies were quite diverses, varying in diameter from 4-6 cm; Color of the colonies is varied with major colors such as bright yellow, dark yellow, dark green, blue gray, black, some white spongy colonies. The number of molds ranges from 104 to 106 cells/mL. With the same samples collected on the market, the isolates obtained were mostly bacterial, with very few fungi grown and grown on isolated media. After separating the colonies and purifying, we selected 12 mold varieties representing different color groups for further study.
Conclusion. Fungus are quite diverse mainly in Aspergillus and Penicillium. Of the nine strains of Aspergillus, six species are closely related to Aspergillus flavus and Aspergillus oryzae.
KEYWORDS: Aflatoxine; Aspergillus; soybean sauce; Ban village; Hung Yen.
RÉSUMÉ
Introduction. Les aflatoxines sont des mycotoxines sécrétées par Aspergillus flavus, qui peuvent coloniser dans les voies respiratoires et provoquer une rhinosinusite fongique ou une aspergillose bronchopulmonaire. A. flavus est la deuxième cause d'aspergillose invasive dans le monde.
Objectifs. Les objectifs de l'étude étaient d'évaluer la diversité des moisissures et des toxines fongiques dans certains échantillons de sauce de soja recueillies à Hung Yen qui sont à la cause de problèmes de santé systémique ou respiratoire.
Méthodes. Des échantillons ont été prélevés au hasard dans certaines familles avec une production traditionnelle de sauce de soja et certains échantillons ont été vendus sur le marché dans le village de Ban - ville de Hung Yen. Microorganismes: fongique isolé de la sauce de soja recueillie dans le village de Ban - ville de Hung Yen.
Résultats. Le résultat a montré que les colonies fongiques étaient assez diverses, leur diamètre variant de 4 à 6 cm. La couleur des colonies varie avec les couleurs principales telles que le jaune vif, le jaune foncé, le vert foncé, le gris bleu, le noir, certaines colonies blanches spongieuses. Le nombre de moules varie de 104 à 106 cellules/mL. Avec les mêmes échantillons collectés sur le marché, les isolats obtenus étaient pour la plupart bactériens, avec très peu de champignons cultivés et cultivés sur des milieux isolés. Après avoir séparé les colonies et purifié, nous avons sélectionné 12 variétés de moisissures représentant différents groupes de couleurs pour une étude ultérieure.
Conclusion. Les champignons sont assez divers principalement de la famille d’Aspergillus et Penicillium. Parmi les neuf souches d'Aspergillus, six espèces sont étroitement apparentées à Aspergillus flavus et Aspergillus oryzae.
MOTS CLÉS: Aflatoxine; Aspergillus; sauce de soyage; Ban village; Hung Yen.
INTRODUCTION
Aflatoxins are mycotoxins secreted by Aspergillus flavus, which can colonize the respiratory tract and cause fungal rhinosinusitis or bronchopulmonary aspergillosis. A. flavus is the second leading cause of invasive aspergillosis worldwide. Inhalation of aflatoxins has been associated with occupations involving exposure to environmental molds24, such as grain processing. However, the effects of inhaled aflatoxins or aflatoxin-producing fungi on the respiratory tract are not well characterized. There is some evidence that airway cells can activate aflatoxins in vitro4,25 and in vivo26,27,28, though the link between aflatoxin exposure and human lung cancer is unclear. Aflatoxin is a toxin produced by several species of Aspergillus fungi, such as A. flavus, A. parasiticus, and A. nomius (Payne and Brown, 1998). These fungi usually grow in tropical areas where the air is high. They often infect cereals like rice, corn, peanuts, beans before or after harvest. There are some major mycotoxins that are B1, B2, G1, and G2; where B1 is the most common type found in nature. When people and animals eat foods containing aflatoxin, health may be affected because this toxin can damage the central nervous system, heart, lungs, cause cancer, inhibit the immune system. translate or affect the structure of DNA. Genetic defects can cause fetal malformations, or abortion (Kumar et al., 2017). Soybean sauce has been used in the late 19th century and it has become one of Vietnam's specialty dishes. There are many soybean sauce homelands especially in Ban town at Yen Nhan village, My Hao district, and Hung Yen province. This is a village known as a famous soybean sauce for making long-standing. Soybean sauce is the indispensable spice when eating foods such as cake bread, rice crackers, water spinach, salt eggplant ... or used as spices to caramelize fish or meat. Bean sauces are used by many people everywhere in Viet Nam and in every kitchen of the family. Soybean sauce is a nutrient-rich product. It is made in the traditional way (incubated in jars) and used for molding, the scientists are concerned about the composition of soybean fungus such as Aspergillus oryzae (fungus mainly used to ferment soybean) and mixed with other fungi, such as Aspergillus flavur.
To clarify the issues mentioned above, we decided to carry out the project: "Research evaluates the diversity of mold and their toxins in the samples of the soybean sauce".
OBJECTIVES
Objectives of the study was to evaluate the diversity of mold and fungal toxins in some soybean sauce samples collected in Hung Yen.
METHODS
Study Materials
Soybean sauce samples
Samples were randomly collected in some families with traditional soybean sauce production and some samples were sold on the market in Ban village - Hung Yen town. Microorganisms: fungal isolated from the soybean sauce collected in Ban village - Hung Yen town. The study was conducted at the Laboratory of Biotechnology - Microbiology, Hanoi University of Pedagogy from June 2017 to April 2018.
Chemical products
Sucrose, NaNO3, KH2PO4, MgSO4, KCl, FeSO4, SDS, CH3COONa, agarose, manufactured by Merck. The medium used for isolation and culture of microorganisms (Czapex-Dox Environment) contains the following compound: 30g Saccharose; 3g NaNO3; 1g of K2HPO4; 0.5 g MgSO4; 0.01g FeSO4; 20 g of jelly; 1000 ml distilled water; pH = 5.5.
Equipments
Heraeus - Germany, Heraeus - Germany, BR300LF (Japan) and Memmert (Germany), Centrifuge (Sorwall super T21 - USA), Micropipette (Gilson - France), pH meter (Swiss MP200R), Analyzer scale (Precisa XT 320M - Switzerland) - Instruments needed at Biotechnology Laboratory - Microbiology. Box, triangular vase 100 ml, test tube, eppendoff, measuring cup, glass funnel.
Research methods
Sampling method
Samples were taken randomly in some families with traditional soybean sauce production and some samples were sold on the market in Ban village - Hung Yen town. The samples were stocked in clean boxes, others were intact, numbered and transported to the laboratory for isolation.
Method of isolation
Isolation of samples obtained on Crapek-Dox medium with the use of sterile equipment. One ml of each sample was injected into a test tube containing 9 ml of physiological saline (0.9% NaCl), sterile, and shaken and diluted to 10-5. Vacuum 0.1ml of diluted solution at 10-3, 10-4 ,10-5 in small concentrations on the surface of prepared Crapek medium in different cage boxes. Samples were translated on the surface of the environment, raised in a refrigerator at 30oC. After 3-4 days, it was taken out the cage boxes for counting the number of moldy colonies and selecting the separate mold colonies to transplant.
Method of preservation and use of germs
Sterile needle was used to separate the mold from the colonies separately to the prepared Czapek-Dox agar slant tube. After 3-4 days of culture at 30oC it was removed for inspection, removal of the contaminated tubing after separation of the mold to other tubes, finally obtained pure germs.
Method of observing mold morphology
Slag method: The molds were placed in the Czapek-Dox medium and then packed in a cage box. Sterile knife was used to cut a slit, put a glass slides on the gully just slit. Covered the cage box, keeping it in the warmer 30 oC for 3-4 days. Then, taken a look under the microscope at magnifications of 10x, 40x. Agar block method: Prepared a sterile Czapek-Dox environment in a cage box to form a thin layer. To dry the environment, drill the cork drills into jar blocks. Placed the mushrooms on the prepared sterile slides, then strained the fungus on the newly formed blister. Then, covered the lamen, placed the slides in sterile cages, keeped in the warmer at 30oC for 3-4 days. Mold specimens were observed under a microscope at magnifications of 10x, 40x.
Method of collecting bioblock and DNA extraction
Bioblock culture: Prepared a liquid Crapek-Dox medium, and added to each 100-mL flask a volume of 20-25 ml of sterile medium. Mold was cultived into triangular pots. Raised the shaker at 30oC at 150 rpm for 2-3 days. Cultivation of mold bioblock was given into 2 ml sterile eppendorf tubes. Centrifugated at 13000 rpm, removed of environmental fluid, washed of bioblock obtained by sterile distilled water. Storaged bioblock was obtained in a refrigerator at -20°C.
Total DNA separation method
Added liquid nitrogen to the 2 ml eppendorf tube containing mold bioblock using sterile plastic rods crushed bioblock; Added 600μl SDS buffer to sample tubes, shaked well, incubateb at 70oC for 30 minutes, then switched to -70oC for 15 minutes and then switched back to 70oC for 15 minutes; Adding 300 μl 3M CH3COONa (pH 5.2), centrifugated at 13,000 rpm for 20 minutes at 4° C. Aspirated switch to a new sterile eppendoff tube; Added isopropanol at a 1:1 ratio to precipitate DNA.
Centrifugated at 13,000 rpm for 5 minutes at 4° C to precipitate DNA; Washed DNA precipitate with 500 μl of 70% ethanol, and dry precipitate by allowing the ethanol to evaporate completely in a sterile culture box; Add 50 μl TE buffer solution (Tris-EDTA, pH8) to the DNA sample.
Total electrophoresis of DNA was done on gel agar plates.
PCR method
Amplification of the 18S rDNA gene sequence by PCR using primers: ITS4 (5'-CCTCCGCTTATTGATATGC-3 ') and: ITS5 (5'-GGAAGTAAAAGTCGTAACAAGG-3'). Heat cycle for reaction: denaturation at 95°C for 3 minutes; Repeated 35 cycles: 95°C Heat cycle for reaction: denaturation at 95°C for 3 minutes; Repeat 35 cycles: 95°C for 1 minute, 52°C for 30 seconds and 72°C for 1 minute; 72°C for 10 minutes and 4°C for product storage. The DNA sequence obtained was compared with the gene sequences in the gene bank to determine the degree of similarity with the sequences available in the gene bank. MEGA 6.06 software was used to compare nucleotide sequences and plant genotypes of selected strains.
Electrophoretic method
Weighed 0.3 g of agarose into a triangular flask containing 30 ml of TAE solution; Boiled to completely dissolve agarose by microwave; Allowed to cool to 65°C and gave 1.5 μl of DNA-staining solution (redsafe 20000 ×). Pour the agarose mixture into a pre-prepared mold. After 30 minutes, placed the gel in a solution containing TAE solution, draw the comb and look into the wells. Plugged in the power and conduct the test with a voltage of 80 V for 60 minutes. Examined the electrophoresis by scanning and photographing the gel under UV light.
Determination of fungal toxins
Mycotoxins were determined according to the kit of Breen Age company as follows: Weighed the 4 g sample into the 50 ml falcon tube. Added 2 ml of water and 8 ml of ethyl acetate, grinded the sample for 5 minutes, centrifuge 6000 rpm for 5 minutes. Taken 4 ml of surface fluid (ethyl acetate) into a small glass. Used wind to blow out the fluid. Dissolved the dried portion in small cups with 0.4 ml of buffer solution (provided in the kit).
Removed the test strip from the bag and place it on a flat, horizontal surface. Lowly added 4-5 drops of specimen into the specimen pits.
Observed and evaluated the results on the test strips after 5-10 minutes based on the marks on the test strips.
Determination of extracellular enzyme activity
Protease and amylase enzyme activity was determined by the method of drilling holes (agar). Enzyme extracts were extracted from culture medium with 0.01 M acetate buffer, pH 5.5. The active culture medium contains 2% agar and 1% agar (gelatin and casein for protease activity, amylase activity test). Use a 3-hole drilled cork drill on an assay medium (2 holes for enzyme and 1 hole for buffer solution - control) then small 100 μl of enzyme solution. Store in a refrigerator 4°C for 6 hours and then keep it in the warmer 37°C for 24 hours. Enzyme activity was examined by staining solution: using an iodine solution to test for amylase activity and using saturated (NH4)2SO4 solution to test for protease activity.
RESULTS
Results of isolation
When observing Petri dishes after incubation at 30°C for 3 days, the result showed that the moldy colonies were quite diverses, varying in diameter from 4-6 cm; Color of the colonies is varied with major colors such as bright yellow, dark yellow, dark green, blue gray, black, some white spongy colonies. The number of molds ranges from 104 to 106 cells/mL. With the same samples collected on the market, the isolates obtained were mostly bacterial, with very few fungi grown and grown on isolated media. After separating the colonies and purifying, we selected 12 mold varieties representing different color groups for further study.
Characterization of colony morphology and spore organism of selected strains of mold
We have cultivated selective molds on the Czapek-Dox environment. After about 3 days of incubation at 30oC, observe and describe the morphology and size of the colony, observe the reproductive organs under the microscope. The results showed that the mold collected from the specimen was not only characteristic of Aspergillus genus, but also some strains of Penicillium genus. Of the 12 strains we studied, 9 species were characterized by Aspergillus genus and 3 strains were characteristic of Penicillium genus.
The strains are characterized by Aspergillus
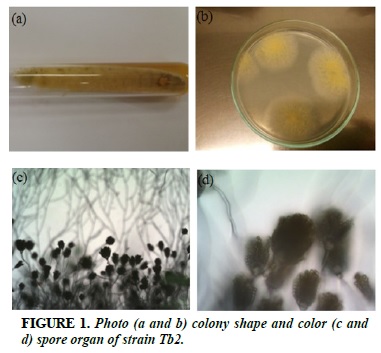
Tb2
White strands of fibrils, when sporulated, colonies turn yellow (Figure 1a and b). The filamentous, branched spherical filament of the sporangium, which radiates around the sporangium (Figure 1c and d).
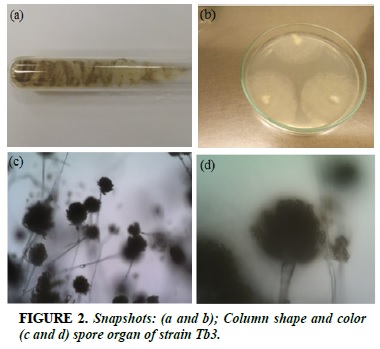
Tb3
White strands of fibrils, when formed spores, colonies turn yellow green (Figure 2a and b). Congenital biopsy, long sporulation, enlarged bulge, sporadically arranged sperm surrounding the spherical pouch (Figure 2c and b).
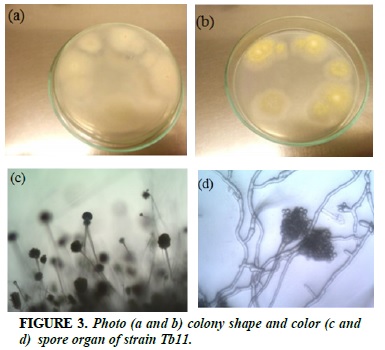
Tb11
White strands of fibrils, when sporulated, colonies turn yellow (Figure 3a and b). Branched dorsal, mesentery-shaped vesicles, sporangium arranged into radial ray masses (Figure 3c and d).
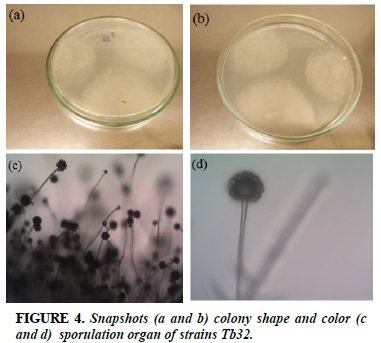
Tb32
White strands of fibrils, when formed spores, colonies turn to light yellow (Figure 4a and b). Sporophore spp., Spherical top, spherical, at the top of the bladder with 1 to 2 layered, radial spheres (Figure 4c and d).
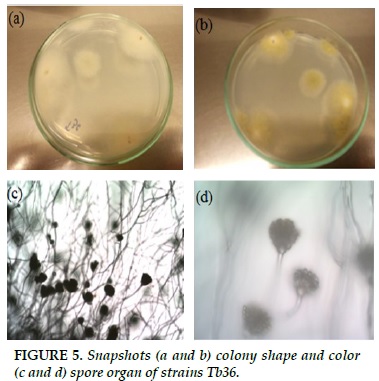
Tb36
White strands of fibrils, when formed spores, colonies turn yellow green (Figure 5a and b). Peak, spherical, sporangially arranged spores about 2-5 spores are attached to a broad, broad sphincter (Figure 5c and d).
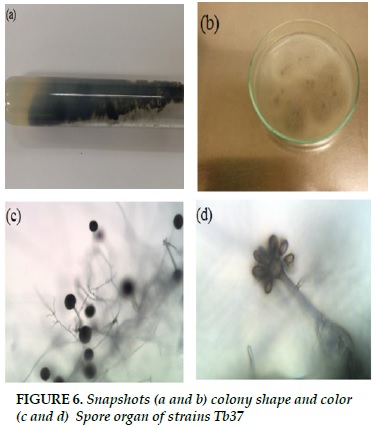
Tb37
Black fibrous strands, when spores formed, colonies turn black (Figure 6a and b). Peak, spherical, spherical spheres arranged directly into the puff (Figure 6c and d).
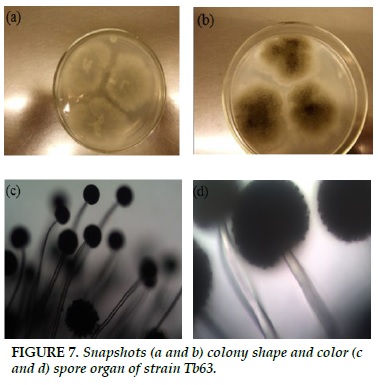
Tb63
White striatum, when spores formed, turned to black (Figure 7a and b). Non-branched sporangium, mesentery, sporangium at the top of the bladder (Figure 7c and d).
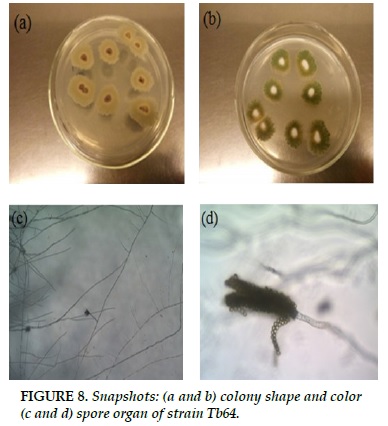
Tb64
White strands of fibrils, when formed spores, colonies turn blue (Figure 8a and b). The branched branches, spores attached together form a long chain, forming long chains (Figure 8c and d).
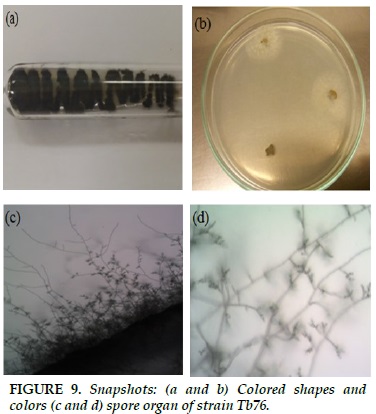
Tb76
Blue-black fibrous structure, in sporangium formation, colonies turn to dark blue (Figure 9a and b). Short, branched gas generator. Spherical, parallel or stringed (Figure 9c and d).
The strains are characteristic of Penicillium genus
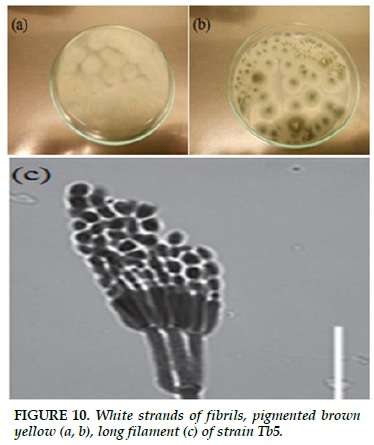
Tb5
White strands of fibrils, when formed spores, colonies turn blue, pigmented brown yellow (Figure 10a and b). The short-lived, short-haired gas sprout, which binds and binds like a broom, sporulates into a long filament (Figure 10c).
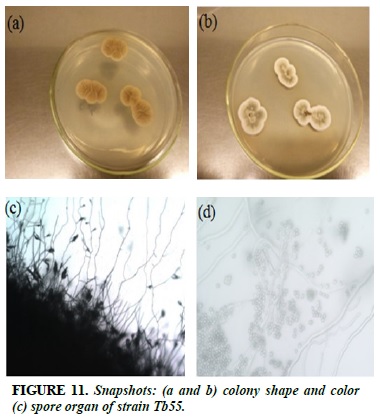
Tb55
The brown substrate, when spores form, turns gray (Figure 11a and b). Branches, elliptic spores, bushes around the spore, branched spores, elongated.
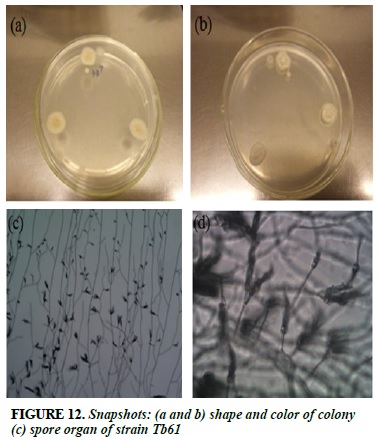
Race Tb61
White strands of fibrils, when sporulated, colonies turn blue gray (Figure 12a and b). The branched branched, spore-shaped spore is arranged in the form of a broom (Figure 12c and d).
Results of strain selection measured by biomolecular techniques
We cultivated and extracted total DNA of 13 strains of mold. The results shown in Figure 16 show that we have successfully separated the total DNA of selected molds.
We used total extracted DNA samples to make a 18S rDNA gene amplified by ITS4 and ITS5 primers (see method section). After the PCR reaction, we conducted the product test by electrophoresis. The results show that we have obtained clean PCR products in the range of 550-600 bp.
The product of the PCR reaction was sent to 1st Base (Singapore) for gene sequencing. The sequence of genes we have used is comparable to the homology sequences found in the gene pool of the world. Comparison results were used to construct the species based on the Mega 6.06 software. Sequences of 12 strains were selected from among 9 strains of Aspergillus and 3 strains belonging to genus Penicillium. Of the 9 strains of Aspergillus, 6 species (Aspergillus sp. Tb2, Aspergillus sp. Tb3, Aspergillus sp. Tb32, Aspergillus sp. Tb36 and Aspergillus sp. Tb37) have sequences similar to the genomes of Aspergillus flavus and Aspergillus oryzae species found in the World Genebank with a homology level of 99 to 100%. Two strains (Aspergillus sp. Tb11 and Aspergillus sp. Tb37) had a homologous 100% sequencing sequence with Aspergillus flavus EFB01. The other three Aspergillus strains have similar levels to other species: Aspergillus sp. Tb64 is similar to Aspergillus unguis DTO 325-C2, Aspergillus sp. Tb76 is similar to Aspergillus niger (99.8%), Aspergillus sp. Tb63 is similar to Aspergillus tubingensis strain CNU081066 (99.8%).
Three strains of Penicillium have similar levels of 18S rDNA gene sequences with different species: Penicillium sp. Tb55 is similar to Penicillium chrysogenum 108 (99.6%), Penicillium sp. Tb5 is similar to Penicillium oxalicum 1034 (99.6%), and Penicillium sp. Tb61 is similar to Penicillium citrinum S115 (99.6%).
Determination of Aflatoxin in samples and estimation of Aflatoxin feasibility of selected strains
We tested the total Aflatoxin content in the samples by means of the total Aflatoxin determination kit of GreenAge. The results showed no fungal toxins detected in the samples. Aspergillus sp. Tb2, Aspergillus sp. Tb3, Aspergillus sp. Tb11, Aspergillus sp. Tb32, Aspergillus sp. Tb36 and Aspergillus spp. Tb37) were cultured in close proximity to the strains. of Aspergillus flavus and Aspergillus oryzae on sticky rice to form mold - similar to the first step in the process of making soy sauce. After 1 week of culture, we extracted and identified the total Aflatoxin. Results showed that Aspergillus sp. Tb37 has an Aflatoxin-positive. Thus, although aflatoxin has not been identified in the sample for use in the study, the results of this study suggest that there are strains capable of producing aflatoxin in these samples.
Research on the selection of strains of mold for safe production
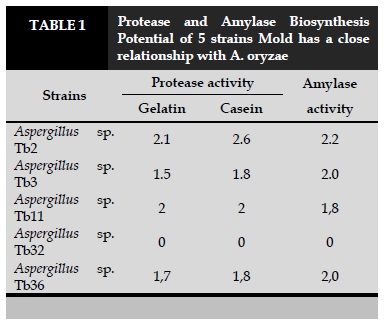
We tested extracellular enzymes of 5 strains (Aspergillus sp. Tb2, Aspergillus sp. Tb3, Aspergillus sp. Tb11, Aspergillus sp. Tb32 and Aspergillus sp. Tb36) which are closely related to A. oryzae and do not produce Aflatoxin. The results are presented in Table 1.
Results showed that only Aspergillus sp. Tb32 is not capable of biosynthesis of protease and amylase, the remaining four are capable of biosynthesis of the two enzymes mentioned above with different strengths (Table 1). Aspergillus sp. Tb2 is capable of biosynthesis of both enzymes is quite high, and the results describe the morphology and comparison of ITS gene sequence shows that this strain is closely related to A. oryzae species, so We have chosen this strain to keep the breed to orient in safe production.
CONCLUSION
We isolate and evaluate the diversity of molds in the samples collected. Fungus are quite diverse mainly in Aspergillus and Penicillium.
Of the nine strains of Aspergillus, six species are closely related to Aspergillus flavus and Aspergillus oryzae.
Results of Aflatoxin screening showed that the samples were in aflatoxin-negative study, but in the isolates were 1 strain (Aspergillus sp. Tb37) with positive Aflatoxin expression.
Protease and amylase biosynthesis of 5 non-toxic strains and their close association with Aspergillus oryzae were evaluated. Results showed that 4/5 strains were capable of biosynthesis of both enzymes, of which Aspergillus sp. Tb2 has the potential to produce powerful enzymes that have been selected and maintained for safe production.
New contributions of the topic
The study assessed the diversity of mold in some soybean samples.
The study evaluated the content of aflatoxin toxin in the samples used in the study. It also evaluates the ability of aflatoxin formation for six molds closely related to Aspergillus oryzae and Aspergillus flavus.
This study contributes to producers and consumers of mildew, fungal toxin aflatoxin content, as well as the risk of fungal toxin formation.
This study selected the non-toxin-producing Aflatoxin, which at the same time produced strong protease and amylase biosynthesis in order to guide the development of safe production.
CONCFLICT OF INTERREST
Non.
REFERENCES
1. Kumar P, Mahato DK, Kamle M, Mohanta TK, Kang SG. Aflatoxin: A global concern for food safety, human health and their management. Frontiers in Microbiology. 2017; 7: 2170.
2. Payne GA, Brown MP. Genetics and physiology of aflatoxin biosynthesis. Annual Review of Phytopathology 1998; 36, 329–362.
3. Blackwell D. L., Collins J. G. & Coles R. Summary health statistics for U.S. adults: National Health Interview Survey, 1997. Vital Health Stat 10, 1–109.
4. Chaudhary N. And Marr K. A. Impact of Aspergillus fumigatus in allergic airway diseases. Clinical and translational allergy 1, 4 (2011).
5. Krishnan S., Manavathu E. K. & Chandrasekar P. H. Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses 52, 206–222 (2009).
6. Sutandyo N. Nutritional carcinogenesis. Acta medica Indonesiana 42, 36–42 (2010).
7. Olsen J. H., Dragsted L. & Autrup H. Cancer risk and occupational exposure to aflatoxins in Denmark. British journal of cancer 58, 392–396 (1988).
FIGURES/TABLE
REFERENCES
1. Kumar P, Mahato DK, Kamle M, Mohanta TK, Kang SG. Aflatoxin: A global concern for food safety, human health and their management. Frontiers in Microbiology. 2017; 7: 2170.
2. Payne GA, Brown MP. Genetics and physiology of aflatoxin biosynthesis. Annual Review of Phytopathology 1998; 36, 329–362.
3. Blackwell D. L., Collins J. G. & Coles R. Summary health statistics for U.S. adults: National Health Interview Survey, 1997. Vital Health Stat 10, 1–109.
4. Chaudhary N. And Marr K. A. Impact of Aspergillus fumigatus in allergic airway diseases. Clinical and translational allergy 1, 4 (2011).
5. Krishnan S., Manavathu E. K. & Chandrasekar P. H. Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses 52, 206–222 (2009).
6. Sutandyo N. Nutritional carcinogenesis. Acta medica Indonesiana 42, 36–42 (2010).
7. Olsen J. H., Dragsted L. & Autrup H. Cancer risk and occupational exposure to aflatoxins in Denmark. British journal of cancer 58, 392–396 (1988).
ARTICLE INFO
DOI: 10.12699/jfvpulm.9.26.2018.32
Conflict of Interest
Non
Date of manuscript receiving
14/01/2018
Date of publication after correction
15/4/2018
Article citation
Le Thi Viet H, Lai Duy H, Doan Van T. Study of the diversity of molds and their toxicity for systemic and respiratory health in soybean sauce. J Func Vent Pulm 2018;26(9):32-39.